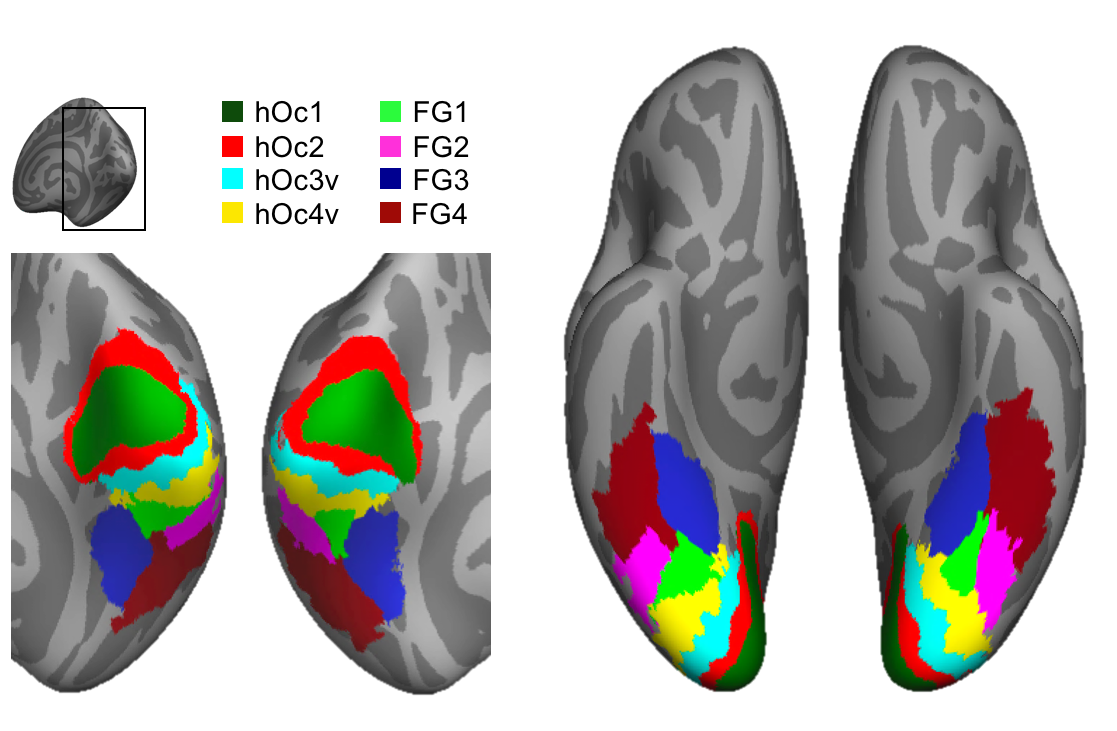
vcAtlas
A cross-validated cytoarchitectonic atlas of the human ventral visual stream.
Developed by the Stanford Vision & Perception Neuroscience Lab.
Atlas regions are available in FreeSurfer as well as BrainVoyager file format:
Download for FreeSurfer BrainVoyager
The cytoarchitectonic atlas is available on the FreeSurfer average surface (fsaverage). Thus your data need to be aligned to the fsaverage brain. Below we describe the files downloaded in the FreeSurfer and BrainVoyager formats.
Citation: Rosenke, M., Weiner, K. S., Barnett, M., Zilles, K., Goebel, R., Grill-Spector, K. (2017). A cross-validated cytoarchitectonic atlas of the human ventral visual stream. NeuroImage, http://dx.doi.org/10.1016/j.neuroimage.2017.02.040
We would like to refer you to our Data in Brief paper as well, we compared two different surface templates for the atlas generation and compared the vcAtlas to the multimodal atlas developed by Glasser et al. (2016): HTML | PDF -->
File and analysis specifics
FreeSurfer
Files
MPM_hemisphere.area.label
|
One label for each cytoarchitectonic area. |
hemisphere.Rosenke_vcAtlas.annot
|
One annotation file per hemisphere, containing all the labels (hOc1 - FG4) for that hemisphere. The annotation file can be loaded o the fsaverage surface like a label file. |
Analysis
Use the cortex-based alignment in FreeSurfer to align your data to the fsaverage brain.
Use the atlas label files to utilize the cytoarchitectonic atlas.
BrainVoyager
The cytoarchitectonic atlas is available in BrainVoyager format on the cortical surface of the fsaverage brain. The files required for cortex-based alignment to the fsaverage cortical surface, as well as the fsaverage cortical surface in BrainVoyager file format are provided with the atlas.
Files
MNI305.vmr
|
Any template in radiological convention can be used to load the fsaverage surface. Here we provide the MNI305 group template. |
fsaverage_hemisphere_SPH_CURVATURE.smp
|
The curvature file of the fsaverage cortical surface. |
fsaverage_hemisphere_SPH.srf
|
fsaverage cortical surface with standardized number of vertices. |
hemisphere_Rosenke_vcAtlas.poi
|
vcAtlas containing all 8 areas: hOc1-FG4; each area is saved as a patch-of-interest (POI). |
Analysis
Please consult the BrainVoyager Support website article on how to run cortex-based alignment. Use the fsaverage cortical surface to utilize the cytoarchitectonic atlas.
Contact
Mona Rosenke rosenke@stanford.edu
Kalanit Grill-Spector kalanit@stanford.edu
Citing
Rosenke, M., Weiner, K. S., Barnett, M., Zilles, K., Goebel, R., Grill-Spector, K. (2017). A cross-validated cytoarchitectonic atlas of the human ventral visual stream. NeuroImage, http://dx.doi.org/10.1016/j.neuroimage.2017.02.040
To acknowledge using this cytoarchitectonic atlas in your research, you might include a sentence like one of the following:
"We used the ventral cROI Atlas developed by Rosenke et al. (2017)..."
"We investigated cytoarchitectonic correspondence using the cROI Atlas of the human ventral visual stream (Rosenke et al., 2017)..."-->